Molecular mechanism regulating transcriptional control of the hig toxin-antitoxin locus of antibiotic‐resistance plasmid Rts1 from Proteus vulgaris.
Dr. Ian Pavelich
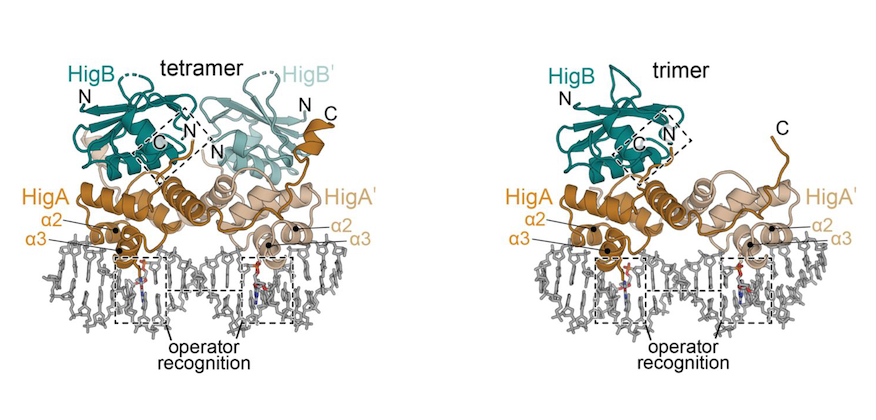
Regulation of ubiquitous bacterial type II toxin-antitoxin (TA) gene pairs occurs via a negative feedback loop whereby their expression is typically responsive to changing levels of toxins at the transcriptional level similar to a molecular rheostat. While this mechanism can explain how certain TA complexes are regulated, accumulating evidence suggests diversity in this regulation. One system for which the negative feedback loop is not well defined is the plasmid-encoded HigBHigA TA pair originally identified in a post-operative infection with antibiotic resistant Proteus vulgaris. In contrast to other type II TA modules, each hig operator functions independently and excess toxin does not contribute to increased transcription in vivo. Structures of two different oligomeric complexes of HigBHigA bound to its operator DNA reveal similar interactions are maintained suggesting plasticity in how hig is repressed. Consistent with this result, molecular dynamic simulations reveal both oligomeric states exhibit similar dynamics. Further, engineering a dedicated trimeric HigBHigA complex does not regulate transcriptional repression. We propose that HigBHigA functions via a simple on/off transcriptional switch regulated by antitoxin proteolysis rather than a molecular rheostat. The present studies thus expand the known diversity of how these abundant bacterial protein pairs are regulated.
- Authors: Ian Pavelich, Marc A. Schureck, Dongxue Wang, Eric D. Hoffer, Michelle Boamah, Nina Onuoha, Stacey J. Miles, C. Denise Okafor, Christine M Dunham.
- https://www.biorxiv.org/content/10.1101/2021.03.04.434028v1.full.